New Target Discovery : CAGE (Cancer Associated GEne)
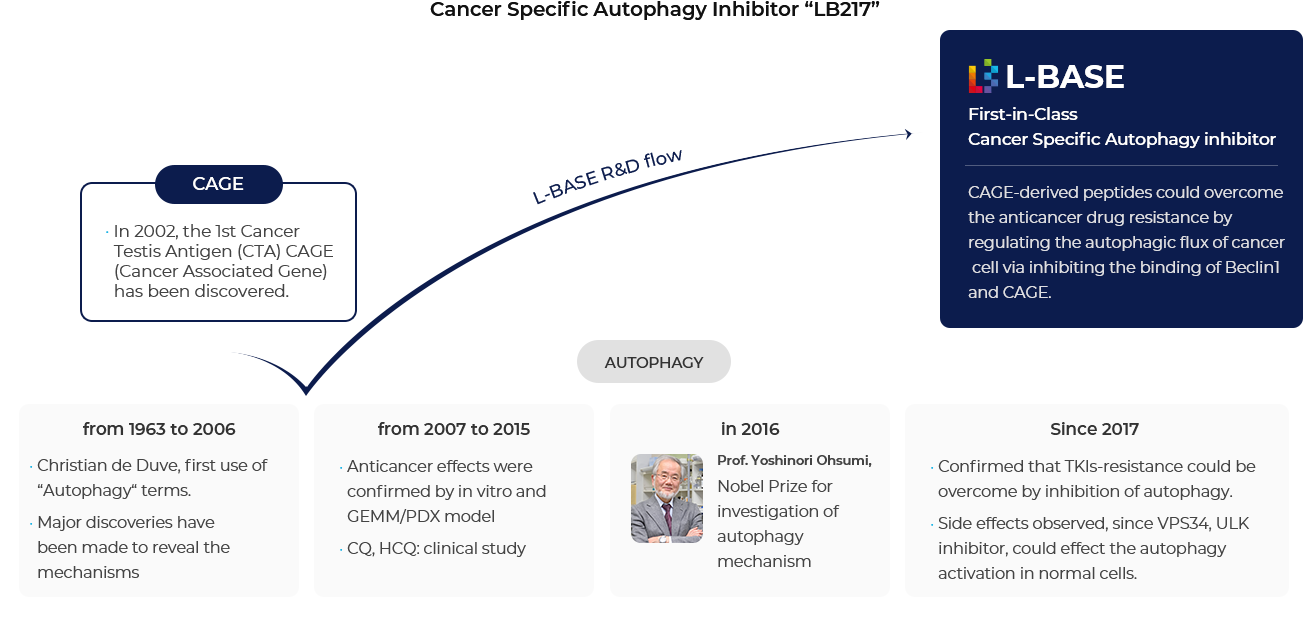
New cancer testis antigen (CTA), CAGE discovered in 2002
CAGE (DDX53, CT26, Cancer Associated Gene), one of the cancer testis antigens (CTA) was discovered first time in 2002.
Since its first discovery in 2002, we recently published 19 SCI papers discovered and confirmed that
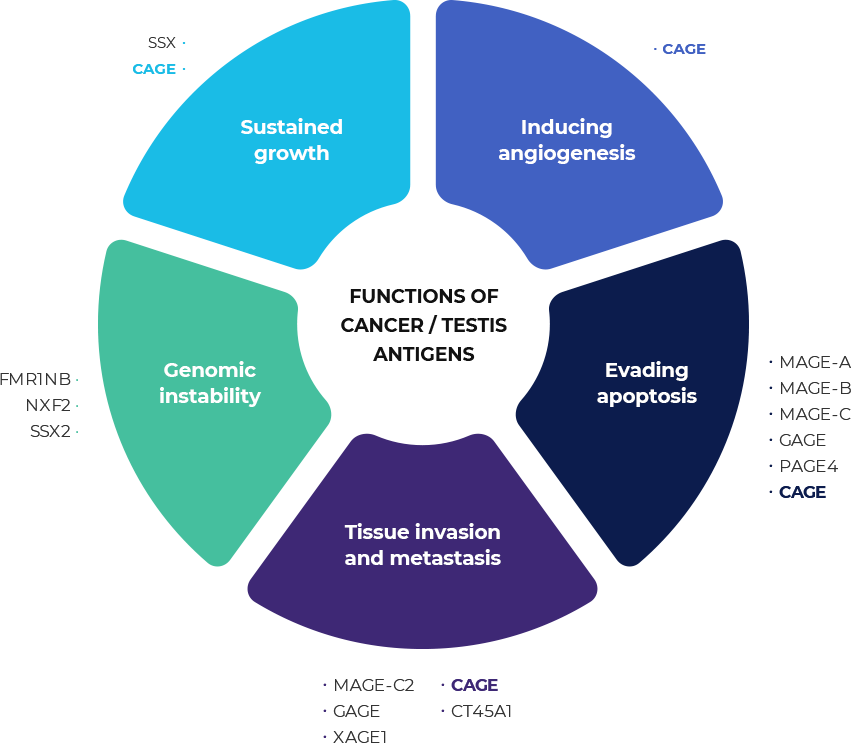
CAGE is related with autophagy activity, anticancer drug resistance, cancer metastasis, angiogenesis induction, cancer stem cell induction
during cancer formation process in several types of cancers.
-
Known as CT26
-
Exhibits anti-cancer activities and overcomes resistance
mutations for existing anti-cancer drugs -
The DDX53 gene is located on the chromosome Xp22.11
-
Molecular weight: 71154 Da
-
CTA (cancer testis antigen) that provides multiple cancer activities and related to EGFR
-
Encoded by the DDX53 gene
-
Size: 631 amino acids
The role of CAGE in cancer formation
CAGE peptide was detected in blood serum of several types of cancer patients, and also was observed in numerous different cancer tissue samples by tissue microarray.
Additionally, it is reported that CAGE binds to Becline1 at early stage of cancer autophagy, and hence
autophagic activity is increased.
It is also reported CAGE is involved in the increase of autophagy activity due to the resistance from the targeted therapy, EGFR TKIs (Tyrosine kinases inhibitors), in non-small cell lung cancer (NSCLC). The expression of CAGE is strongly correlated with EGFR TKIs.
Expression pattern of CAGE in tissue from cancer patients
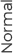

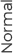

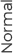

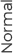

Breast
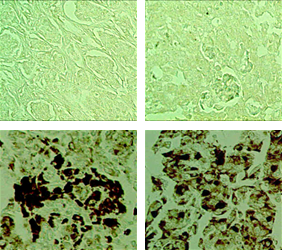
Lung
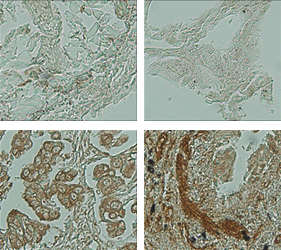
Liver
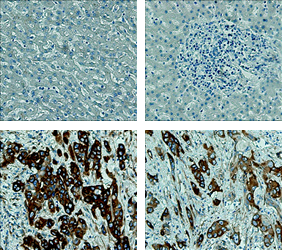
Skin
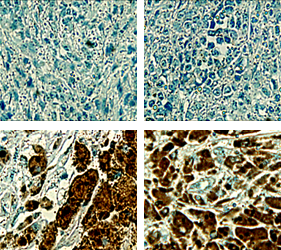
| Tissue | Type | |||
|---|---|---|---|---|
| Normal (adjacent) | Tumor | |||
| Positive (*) | Negative (*) | Positive (*) | Negative (*) | |
| Skin | 0% (0/24) | 100% (24/24) | 19.7% (15/76) | 81% (61/76) |
| Liver | 10% (1/10) | 90% (9/10) | 37.1% (26/70) | 62.9% (44/70) |
| Lung | 12.5% (2/16) | 87.5% (14/16) | 47.6% (40/84) | 52.4% (44/84) |
| Breast | 16.6% (2/12) | 83.4% (10/12) | 52.3% (46/88) | 47.7% (42/88) |
Tissue microarray analysis of the tissue from cancer patients, it is confirmed that CAGE has been overexpressed in breast, lung, liver, skin cancers.
Source: THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 34, pp. 25957–25968, August 20, 2010
* - Number of patients
CAGEpep® Platform for
Anti-cancer Drug R&D
Novel Target
PPI screening
Technology

Indications
CAGEpep® Platform is consists of three main technologies.
-
01
CAGE binding partner
peptide library -
02
Protein-protein interaction (PPI)
inhibitor drug screening system -
03
Protein binding 3D structure modelling based
virtual screening system
CAGEpep® screening system
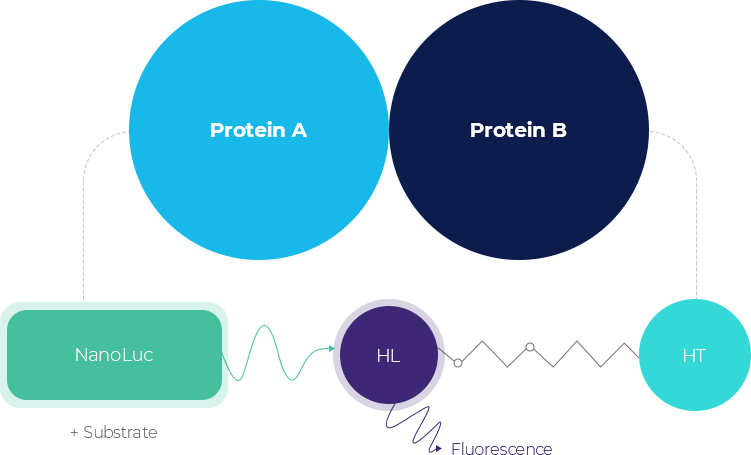
Established the NanoBRET system that utilized to screen the suppression (inhibition) of new candidate drugs on binding effect of proteins with CAGE.
NanoBRET is the analytical method that can determine the specific intracellular PPIs,
it is a technology that determines the intracellular expression of Tag-protein bound CAGE (protein A) and Beclin1 (protein B)
which has higher specificity and better quantification analysis compared to existing FRET method.
Autophagy is?
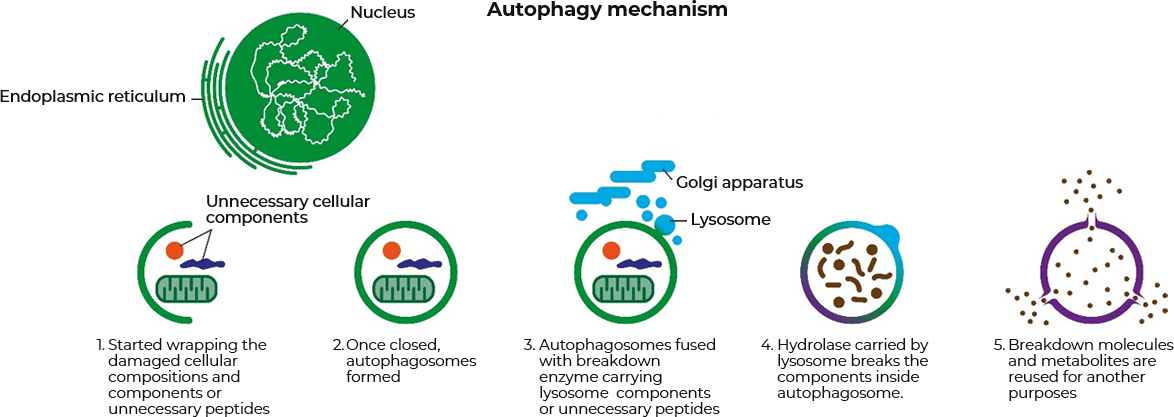
Human body consist of 60 billion cells, and the energy and building blocks required for everyday activities are being created within these cells. During those processes, cell organelle function reduction, protein deformation, cytoplasmic waste occurs. Autophagy is the recycling system for those intracellular wastes and remaining.
Autophagy is originated from Greek words combination of auto-self, and phagy-eat. The first-time autophagy was known through the scientist named Christian du Duve who won Nobel prize by study on cellular structures and functions. And then in 1992, Japanese professor Yoshinori Ohsumi reported the process of autophagy within yeast, a single cell microorganism, and the discovery of 15 related genes later revealed that the autophagy plays an important role in survival. Ohsumi professor has won the physiological medicine in 2016.
Autophagy and Cancer
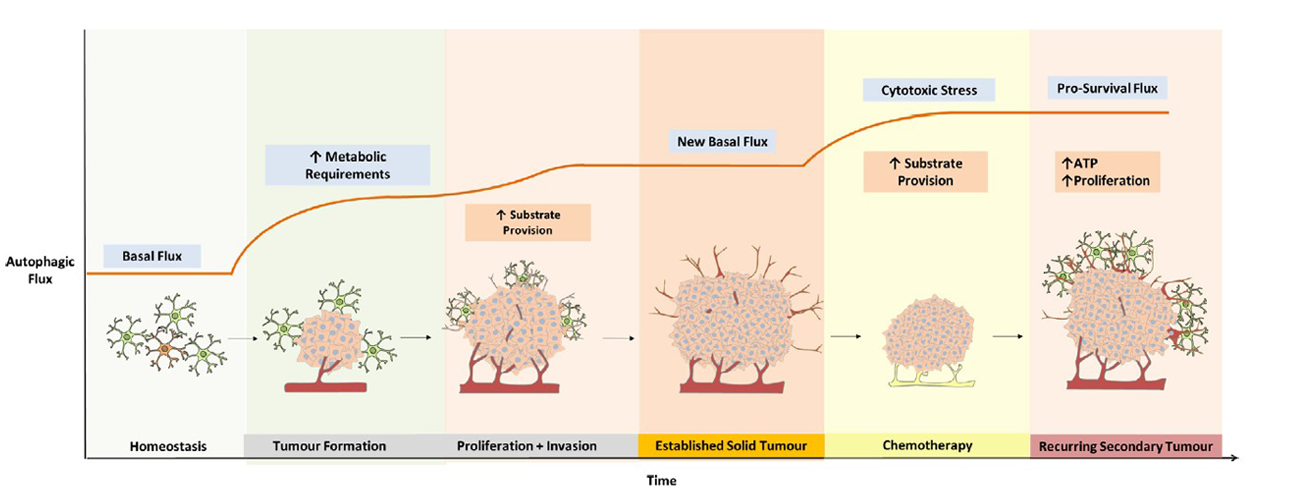 Source: Biochemical Pharmacology Volume 147, January 2018, Pages 170-182
Source: Biochemical Pharmacology Volume 147, January 2018, Pages 170-182
When autophagy does not function properly and accumulate waste matters, it causes numerous disorders.
If residues of peptides are overflown out of the cells, it could cause cancer induced by gene mutation,
If it accumulates in the brain, it could cause the Alzheimer or Parkinson diseases.
Normal cells differentiate into cancer cells, and encountering low-oxygen and malnutrition environment could trigger the stress which results apoptosis
In that condition, autophagy keeps energy supply by recycling waste matters and maintains multiple cell survival mechanisms,
which gives stability to cancer development.
Autophagy and anticancer drug resistance
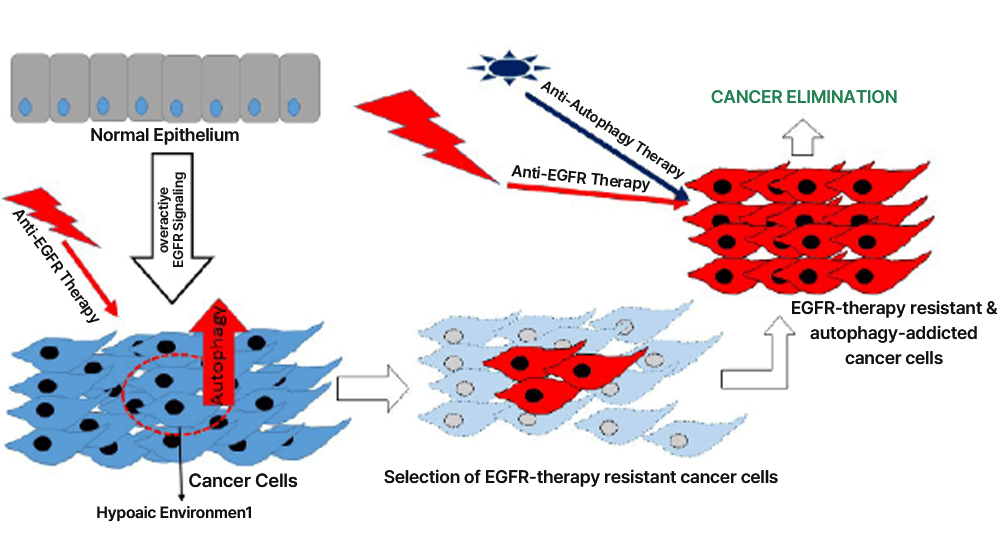 Source: J Adenocarcinoma. 2016, 1(2): 9
Source: J Adenocarcinoma. 2016, 1(2): 9
As an oncogenic factor, autophagy is related to previously existing cancer drug resistance,
which brings a lots of attention from the scientists who developing an anticancer drug.
For example, in case of EGFR TKI (tyrosine kinase inhibitor), a treatment for invasive and metastatic non-small cell lung cancer,
it is reported that the initial treatment was effective, but long-term administration could cause the autophagy-induced resistance.
Autophagy activated during this process is called “protective autophagy”,
and related researches activities are continuing including lung, pancreatic, colorectal, breast, and prostate cancers.
Also, autophagy is closely related to immune response, and inhibition of autophagy could induce anticancer effect by activation of cancer reducing natural killer cells including CD8+,
which gives possibility to combination of autophagy inhibitor and immunotherapy drugs for the patients with immunotherapy drug resistance.
Differentialtion strategy of L-Base
“Maintain the autophagy in the normal cells, only inhibit the autophagy in the cancer cells.”
After all, since the intracellular substances involved in autophagy are the same for both normal and cancer cells,
the key for the success or failure of the cancer treatment is the autophagy inhibition because it works only in cancer cells and does not work in normal cells.
Therefore, a technology precisely targeting this cytoprotective autophagy is emerging as a crucial issue.
Main point is the finding the switch that can selectively turn off the autophagy only in cancer cells.
If we find and control this crucial switch, development of the cancer specific autophagy inhibitor would be possible.
L-Base is developing the new drug that inhibits the specific peptides that have important role in cancer cell maturation, metastasis, immune evasion.
Currently existing new drugs in L-Base pipelines are based on the mechanism of the autophagy inhibitors through PPI inhibition.
Our forerunning candidate, developed for the treatment of non-small cell lung cancer,
is the innovative new candidate drug with the ability to differentiate the cytotoxic autophagy and cytoprotective autophagy,
which is the main issue for every company who is targeting the autophagy as a target.